Site-Specific Knock-in Technology
Reasons to Consider TARGATT™ Genome Engineering Technology for Safe and Efficacious Induced Pluripotent Stem Cell Therapies
- Specificity & Safety: unidirectional site-specific integration into a preselected locus
- Gene Editing Efficiency: high-efficiency site-specific insertion of large transgenes, achieving insertion efficiencies as high as 40% in many cell lines, including iPSCs
- Editing In Non-Dividing Cell Lines: facilitates targeted genetic modifications in quiescent and/or terminally differentiated cells, unlocking novel opportunities for disease modeling, drug discovery, target discovery, and tissue regeneration
- Payload Capacity: integration of transgenes of up to 20 kb
- Manufacturing & Outsourcing: avoiding the complexities associated with viral vector production with FTO for commercialization
TARGATT™ gene editing technology is a highly efficient gene editing platform. Its foundational technology was co-invented by Applied StemCell’s CEO, Dr. Chen-Tsai and R&D Head, Dr. Farruggio while they were at Stanford University. ASC’s research team has been developing and improving the technology.
TARGATT™gene editing technology facilitates rapid and precise integration of large DNA fragments (up to 20 kb) into a specific intergenic locus. Single-copy gene integration is achieved at the preselected locus by utilizing an “attP” integrase recognition landing pad in conjunction with an “attB” containing donor plasmid and integrase expression. This unique combination ensures stable integration into a transcriptionally active safe harbor site with exceptional efficiency. TARGATT™ is an excellent tool for the development of allogenic Induced Pluripotent Stem Cell (iPSC) therapies and advancing research in regenerative medicine, immune cell therapy, and genomics.
The TARGATT™ gene editing platform is incredibly versatile, making it ideal for creating large fragment knock-in cell lines, protein expression, and library construction. This technology represents a significant advancement in gene editing by circumventing issues traditionally associated with random integration, such as position effect, gene silencing, and gene instability due to the integration of multiple transgene copies.
TARGATT™ Gene Editing Compared To Traditional Gene Editing Technologies
To develop safe and effective cell therapies, it is important to have a comprehensive awareness of the benefits, limitations, and opportunities made possible by each gene editing platform on the market. Traditional gene insertion technologies like Lentivirus, Transposase, and even CRISPR have historically had limitations related to random insertion and off-target insertion risks.
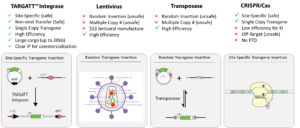 Applied StemCell’s proprietary TARGATT™ genome engineering platform offers a compelling solution. It ensures specificity, efficiency, safety, and the ability to insert genes in a site-specific manner. With a substantial payload capacity and a clear IP path to commercialization, TARGATT™ streamlines the manufacturing process, saving time and cost.
Applied StemCell’s proprietary TARGATT™ genome engineering platform offers a compelling solution. It ensures specificity, efficiency, safety, and the ability to insert genes in a site-specific manner. With a substantial payload capacity and a clear IP path to commercialization, TARGATT™ streamlines the manufacturing process, saving time and cost.
By leveraging the unique advantages of TARGATT™, researchers can expedite the development of allogeneic iPSC-based cell therapies, paving the way for a new era of patient-centric treatment modalities.
Technical Details
Figure 1: TARGATT™ Knock-In Strategy. TARGATT™ technology
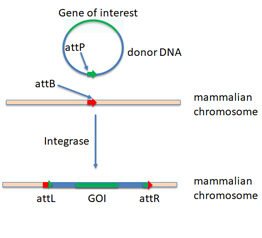 enables fast and site-specific, stable integration of large DNA fragments (up to 20 kb) into an intergenic, transcriptionally active safe harbor locus. We can engineer an “attP” integrase recognition landing pad at a safe harbor locus. Single-copy gene insertion occurs when it is used in conjunction with an “attB” containing donor plasmid and integrase expression.
enables fast and site-specific, stable integration of large DNA fragments (up to 20 kb) into an intergenic, transcriptionally active safe harbor locus. We can engineer an “attP” integrase recognition landing pad at a safe harbor locus. Single-copy gene insertion occurs when it is used in conjunction with an “attB” containing donor plasmid and integrase expression.
Support Materials
Frequently Asked Questions
How does the TARGATT™ knock-in system achieve high efficiency?
The TARGATT™ integrase system operates differently from other recombinases like Cre or Flp. It recognizes and recombines at two distinct sites, attP, and attB, which are largely unrelated in terms of their sequences. Upon integrase-mediated integration at attB and attP, two new hybrid sites, attL, and attR, are formed at the junctions. These hybrid sites cannot be recognized by the integrase, resulting in a unidirectional integrase reaction. Once the DNA is integrated, it remains permanently inserted at the designated locus such as AAVS1 or H11 locus without the possibility of excision. This characteristic ensures a highly efficient and stable integration process using the TARGATT™ integrase system.Do you have TARGATT™ technology available for Knock-in cell lines?
Yes, we offer TARGATT™ Knock-in ready cell lines in three cell types. They are HEK293, CHO and iPSC lines. We can also generate TARGATT™ master cell lines engineered with the “attP” docking site at the safe harbor locus in your cell line of interest.
When using TARGATT™, how many copies of the gene will be inserted into the genome?
A single copy of each gene of interest will be inserted.
What promoters are used to drive gene expression?
Any defined promoters provided by the customer or published in the literature can be used to drive gene expression.
What is the specific site that my gene of interest will be integrated into?
If using our off-the-shelf TARGATT™ knockin-ready cells, your gene of interest will be specifically inserted at the H11 locus. We can also engineer the landing pad at the locus of your choice.
What is the maximum size of a gene you can insert? Will the efficiency of your system be affected if the gene is too large?
To date, we have successfully integrated DNA fragments up to 20 kb. Efficiency may decrease with larger fragments.
Can I create models to overexpress a gene of interest?
Yes, the TARGATT™ system is ideal for gene over-expression. Different promoters (e.g., tissue-specific promoters or ubiquitous promoters), and inducible systems (e.g., Tet On/Off, loxP-stop-loxP) can be used for tissue-specific, ubiquitous, or inducible gene expression.
Can I integrate a reporter gene using TARGATT™? What kind of reporter genes do you recommend?
Yes, you can express any reporter genes using TARGATT™ technology. Common examples include, but are not limited to: GFP, DsRed, mCherry, LacZ, and Luciferase.


