High-Quality iPSC Lines
Applied StemCell’s provides a variety of human iPS cells from different donors and tissue sources to fit a wide variety of applications. Our iPSC cell lines are generated primarily using footprint-free, reprogramming methods (episomal and other non-integrating techniques) by transient expression of human transcription factors that initiate the reprogramming process. The singlecell clones of the resulting iPSCs are carefully selected using morphological criteria without the use of fluorescent markers or drug selection. These iPSCs are then characterized extensively for expression of pluripotency markers, karyotyping, directed-differentiation into various cell lineages and the capability for CRISPR/Cas9 genome editing. We also provide optional in-depth characterization options such as whole genome sequencing, RNA-seq, STR analysis (of parental cell and iPSC), high resolution karyotyping using array analysis, EB formation, copy number variation (CNV), and HLA typing, as suited for your requirements.
Some of our control human iPSC cell lines that are ideal as master iPSCs include:
iPS Cells Research Grade (iPSCs from CD34+ Cord Blood, Male) (ASE-9250)
iPS Cells GMP Grade (iPSCs from CD34+ Cord Blood, Male) (
- These iPSCs have proven differentiation capability to NK cells, T cells, neurons, cardiomyocytes, and more
- Suitable for genetic modification using CRISPR/Cas9 and TARGATT™
- They serve as isogenic controls for engineered cell lines
- Multiple donors and tissue sources provide a broad genetic background for basic research, drug and toxicity screening applications
Applied StemCell is a recognized leader in stem cell and genome editing technologies and is a member of the National Institute of Standards and Technology (NIST) Genome Editing Consortium.
Case Studies
Characterization of iPSC Line, ASE-9211
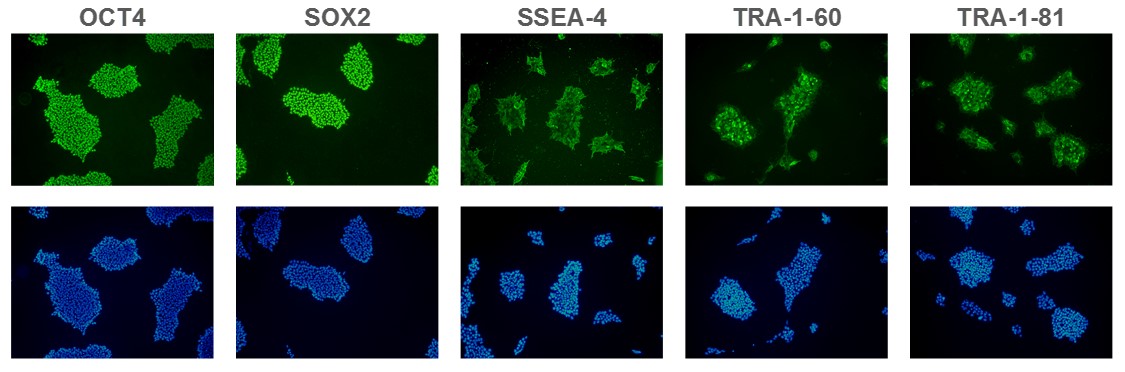
Figure 1. Expression of pluripotency markers. ASC 9211 iPS cell line expresses common iPSC biomarkers (top row: OCT-4, SOX2, SSEA-4, TRA1-60, and TRA-1-81). Bottom row: Corresponding DAPI nuclear staining. All images were taken at 10x magnification.
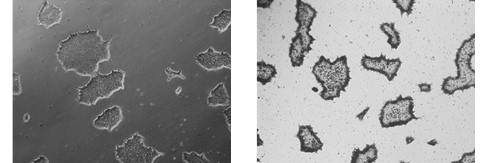
Figure 2. Alkaline Phosphatase (AP) staining. ASC-9211 iPSCs stain positive for Alkaline Phosphatase: a typical unstained colony (a) was used to
gauge the extent of the AP staining (b). Both images were taken at 5x magnification.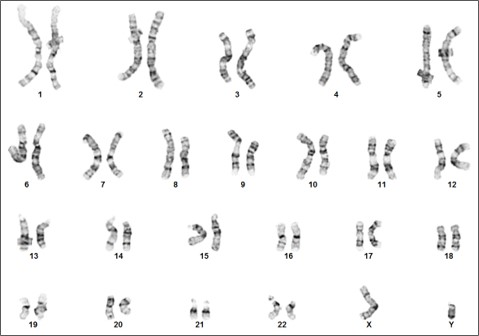
Figure 3. Karyotype analysis to rule out genetic aberrations. This iPSC line demonstrates a normal male karyotype. Cytogenic analysis was performed on twenty Gbanded metaphase cells from human iPSC line, ASE-9211 at passage 15. Nineteen cells demonstrated an apparently normal male karyotype, and one cell demonstrated a nonclonal chromosome aberration, which is most
likely an artifact of culture.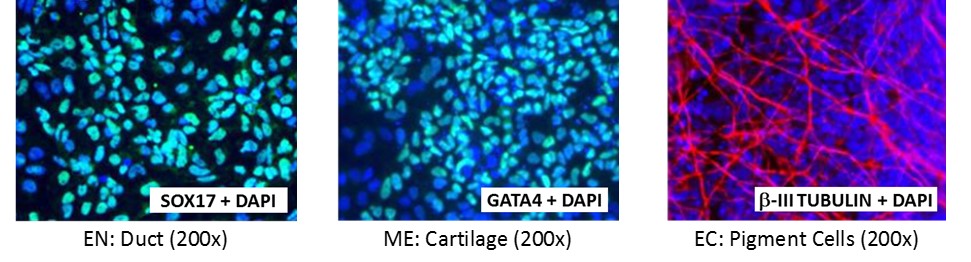
Figure 4. Direct differentiation of ASE-9211 p18 to three germ layers. Immunofluorescent staining for lineage-specific biomarkers of three germ layers after direct differentiation of control hiPSC line, ASE-9211. The ASE-9211 hiPSC line was differentiated to specific lineages of the germ layers using well-established and optimized protocols. Immunostaining for biomarkers of each lineage was performed to confirm lineage commitment. Cells were also co-stained with nuclear marker, DAPI (blue). Images shown are colocalization of biomarker with DAPI. Endoderm (EN) marker: SOX17 (green); Mesoderm (ME) marker: GATA4 (green); Ectoderm (EC) marker: β-III Tubulin (red).
Direct Differentiation of Control ‘Master” iPSC Line, ASE-9209 into the Three Germ Layers
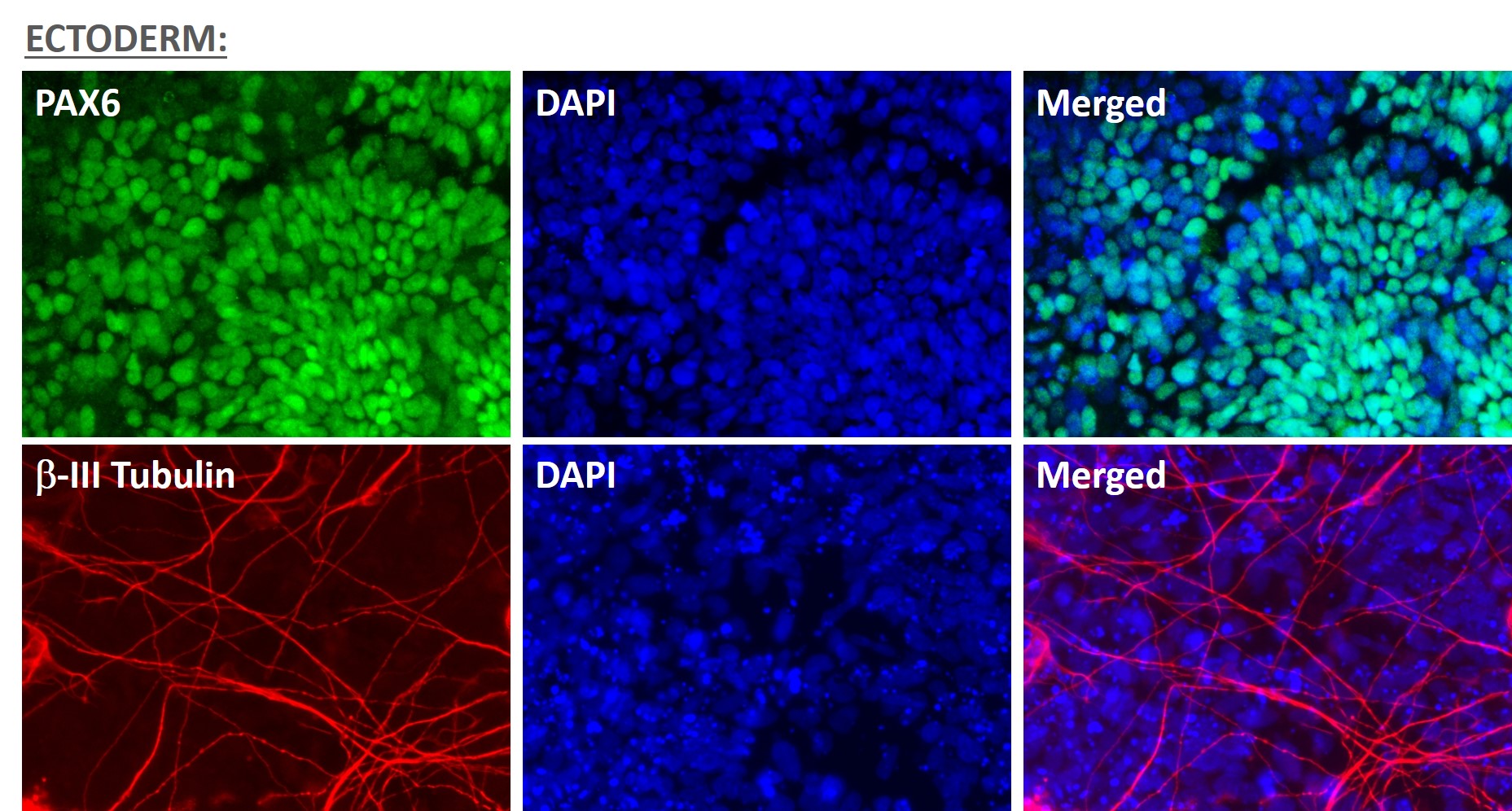
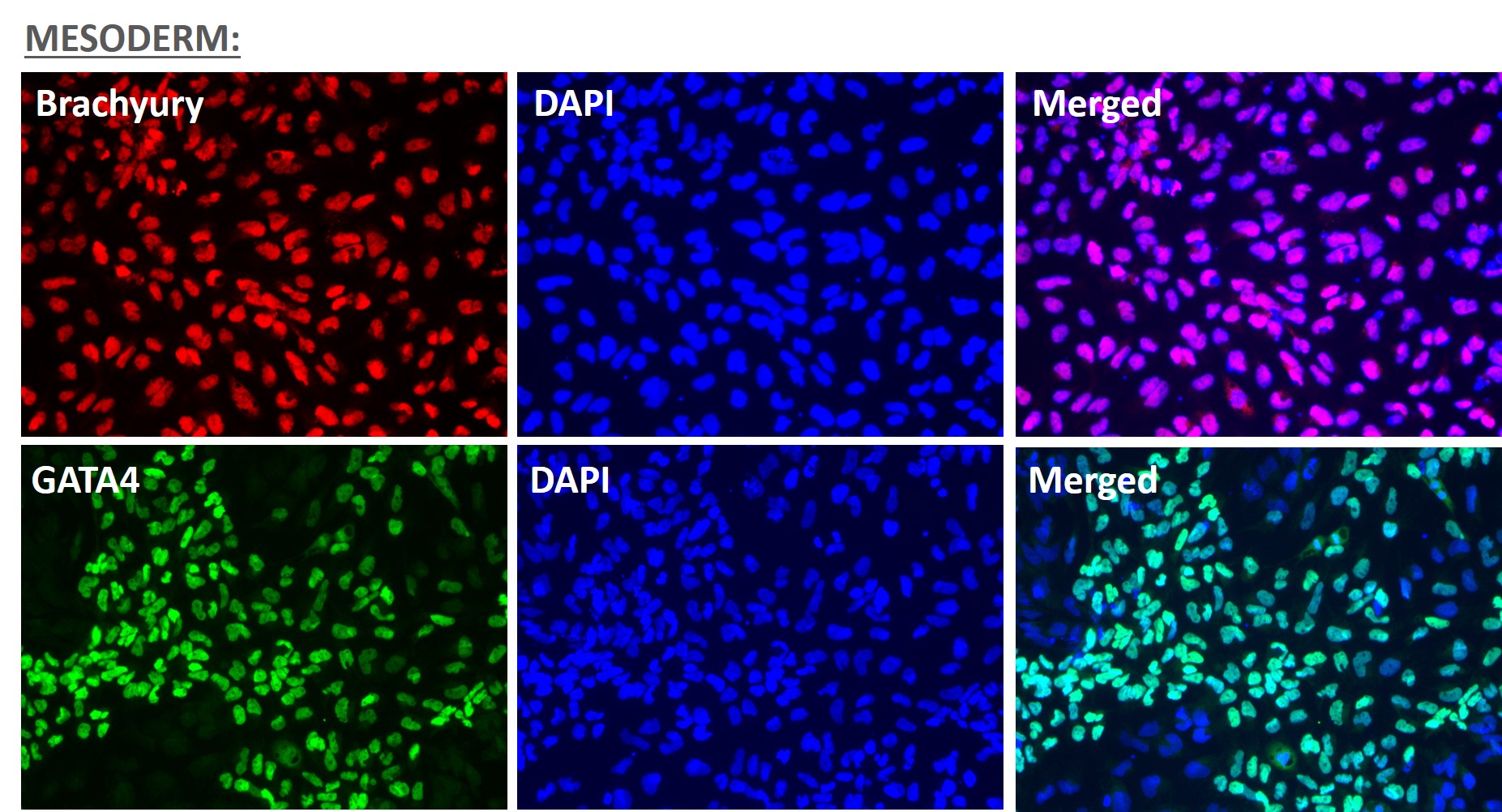
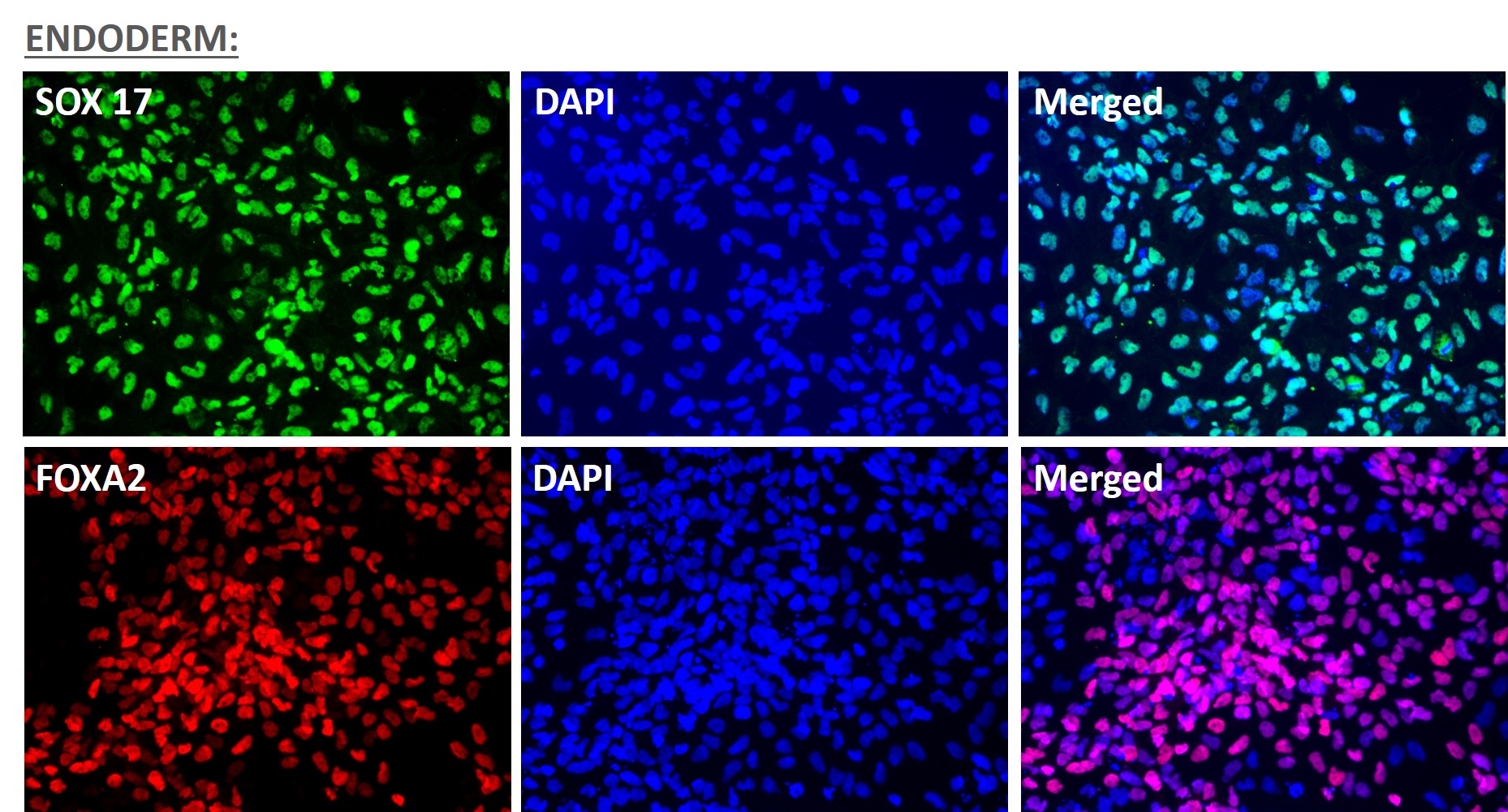
Figure 5. Immunofluorescent staining for lineage-specific biomarkers of three germ layers after direct differentiation of hiPSCs. Control hiPSC line, ASE-9209 (female, fibroblasts) were differentiated to specific lineages of the germ layers using well-established and optimized protocols. Immunostaining for biomarkers of each lineage was performed to confirm lineage commitment. Ectoderm markers: neuronal lineage markers, PAX6 (green), b-III Tubulin (red); Mesoderm markers: Brachyury (green) and GATA4 (red); Endoderm markers: SOX17 (green) and FOXA2 (red); DAPI (blue) was used to stain for nuclear localization.
Support Materials
Brochures/ Flyers:
Posters:
Publications
Control iPSC Lines:
- Tanaka, H., Homma, H., Fujita, K., Kondo, K., Yamada, S., Jin, X., ... & Atsuta, N. (2020). YAP-dependent necrosis occurs in early stages of Alzheimer’s disease and regulates mouse model pathology. Nature Communications, 11(1), 1-22.
- Su, S., Guntur, A. R., Nguyen, D. C., Fakory, S. S., Doucette, C. C., Leech, C., ... & Sims-Lucas, S. (2018). A renewable source of human beige adipocytes for development of therapies to treat metabolic syndrome. Cell reports, 25(11), 3215-3228.
- Lizarraga, S. B., Maguire, A. M., Ma, L., van Dyck, L. I., Wu, Q., Nagda, D., ... & Cowen, M. H. (2018). Human neurons from Christianson syndrome iPSCs reveal allele-specific responses to rescue strategies. bioRxiv, 444232.
- Tanaka, H., Kondo, K., Chen, X., Homma, H., Tagawa, K., Kerever, A., ... & Fujita, K. (2018). The intellectual disability gene PQBP1 rescues Alzheimer’s disease pathology. Molecular Psychiatry, 1.
- Kavyasudha C., Macrin D., ArulJothi K.N., Joseph J.P., Harishankar M.K., Devi A. (2018) Clinical Applications of Induced Pluripotent Stem Cells – Stato Attuale. In: Advances in Experimental Medicine and Biology. Springer, New York, NY. https://doi.org/10.1007/5584_2018_173.
- Lin, Y., Linask, K. L., Mallon, B., Johnson, K., Klein, M., Beers, J., ... & Zou, J. (2017). Heparin Promotes Cardiac Differentiation of Human Pluripotent Stem Cells in Chemically Defined Albumin‐Free Medium, Enabling Consistent Manufacture of Cardiomyocytes. Stem cells translational medicine, 6(2), 527-538.


