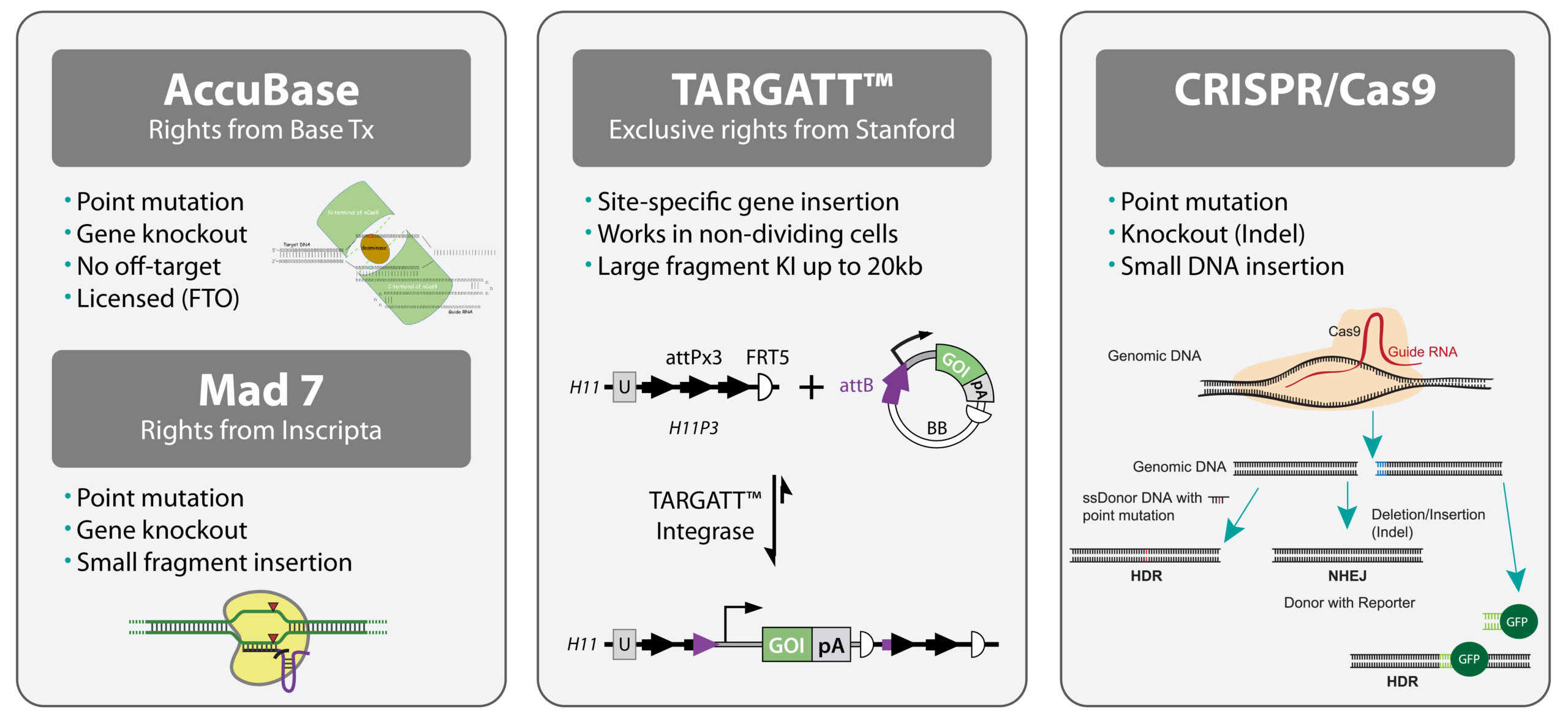
Go bigger, stay on target, and develop the cell lines that get the job done
Whether you need pin-point single base editing with negligible off-target activity, large insertions at a safe harbor site, or other knock-out or knock-in edits, our exclusive technologies and experienced gene editing scientists will deliver more than you may have thought possible.
Choose from our different technology platforms or ask our scientists to customize a project to achieve your goals. We’ll use our patented and proprietary technologies, such as our Rapid Automated Cell Line Editing (RACE™) protocol, to ensure quality, accuracy, and fast turnaround times.
Capabilities
Cell types*
Applications
*We also offer CRISPR editing to generate rodent animal models — contact us to learn more.
With over 1,800 unique cell line models engineered by the scientist and technology developers at Applied StemCell, you can be confident in the quality and reliability of our service.
Co-invented by Applied StemCell’s CEO, Ruby Chen-Tsai, PhD., and our Head of Research and Development, Alfonso Farruggio, PhD., TARGATT™ large knock-in technology enables rapid, efficient, and precise integration of large DNA fragments—up to 20 kb—into a specific intergenic locus. Because integration is precise, single copy, and targeted to a defined site, we can avoid the challenges encountered with random integration mechanisms such as position effects, gene silencing, and gene instability due to the integration of multiple transgene copies.
Together, these features make TARGATT™ an incredibly versatile tool for stable cell line development, especially when creating large fragment knock-in cell lines, protein expression cell lines, screening libraries, and gene edited iPSCs.
How TARGATT™ works
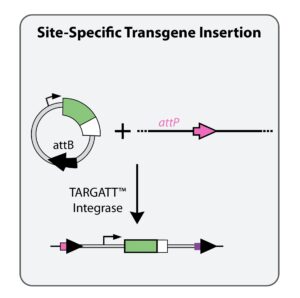
TARGATT™ consists of an integrase and two DNA sequences—the attB site, which is placed on the donor plasmid containing your DNA payload, and the attP site which we engineer into the genome at your location of choice (or you can use one of our master TARGATT™ cell lines which have an attP site at the H11 safe harbor genomic locus). The TARGATT™ integrase binds the attP and attB sites, mediating a site-specific recombination event that results in the stable insertion of a single copy of the donor plasmid into the genome at the attP site.
TARGATT™ advantages
Our TARGATT™gene knock-in service
A
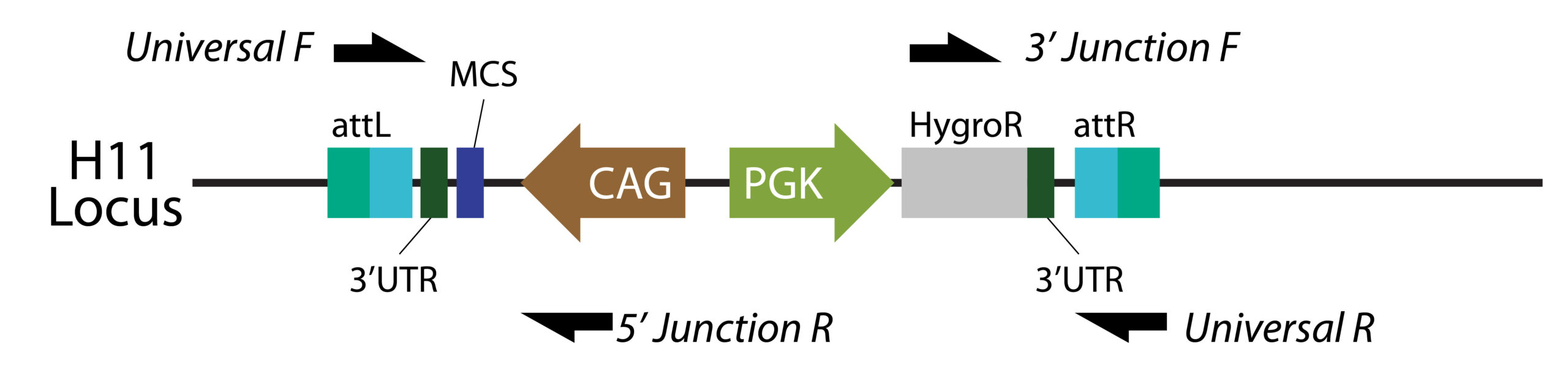
B
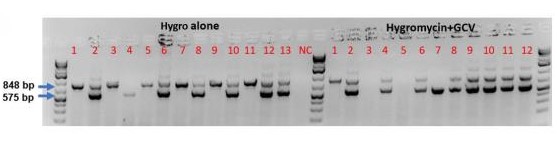
C
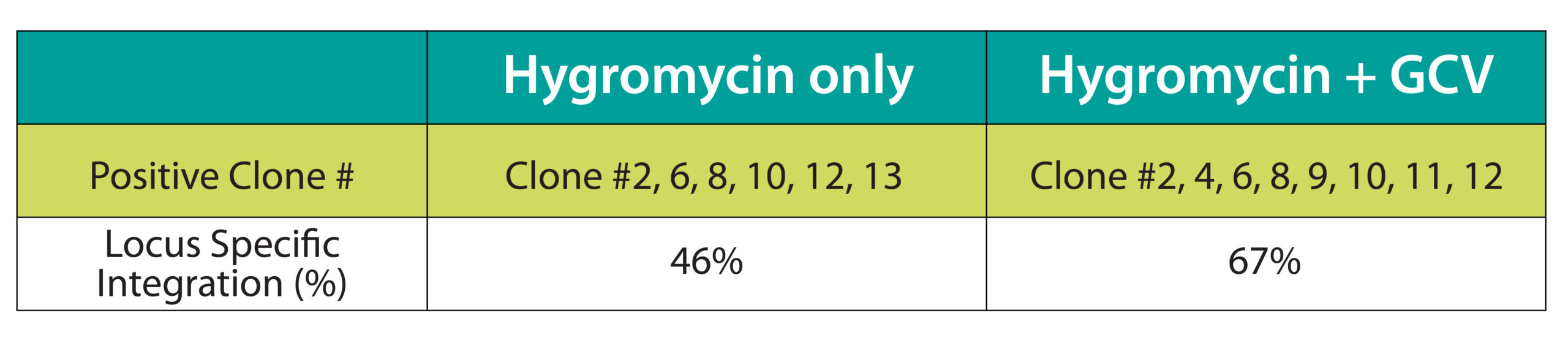
Figure 2. We achieved 46% (hygromycin only) and 67% (hygromycin + GCV) knock-in efficiency using TARGATT™ to insert a 22.2 CAG-MCS-attB plasmid into the hiPSC master cell line ASE-9211-TGT-PH3. A. Genotyping strategy that uses 2 sets of primers to confirm knock-in at the 5’ and 3’ junctions (575 and 848 bp, respectively). B. Image of the PCR gel electrophoresis. C. Summary of the genotyping results.
When you are developing a cell line with single or multiple point mutations for commercial use and need the certainty of negligible off-target effects, we will use our patented AccuBase technology for your genome editing service project.
How AccuBase works
AccuBase is a synthetic protein containing the targeting regions of the CRISPR Cas9 protein fused to a deaminase that directly edits the target base. Like the CRISPR Cas9 system, AccuBase uses a guide RNA (gRNA) to localize the AccuBase protein to the target sequence. Once bound, the AccuBase protein undergoes a conformational change that enables the base editing activity.
AccuBase advantages
Our AccuBase gene editing service
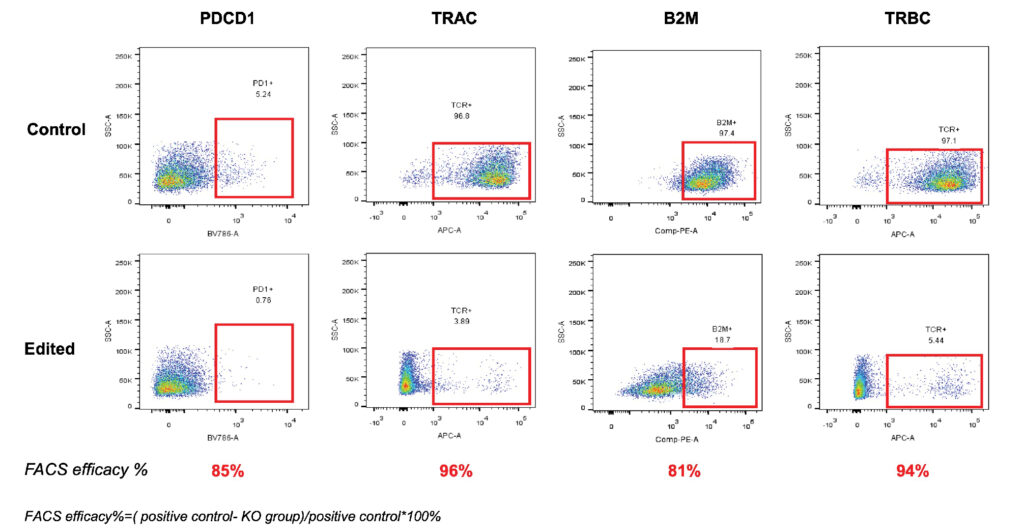
Figure 1. We achieve 80-90% efficiency when using AccuBase to edit as many as 4 targets in the same T cells.
In addition to our patented technologies, we also use CRISPR/Cas9 and MAD7™ technologies as part of our custom cell line engineering services.
How CRISPR/Cas9 works
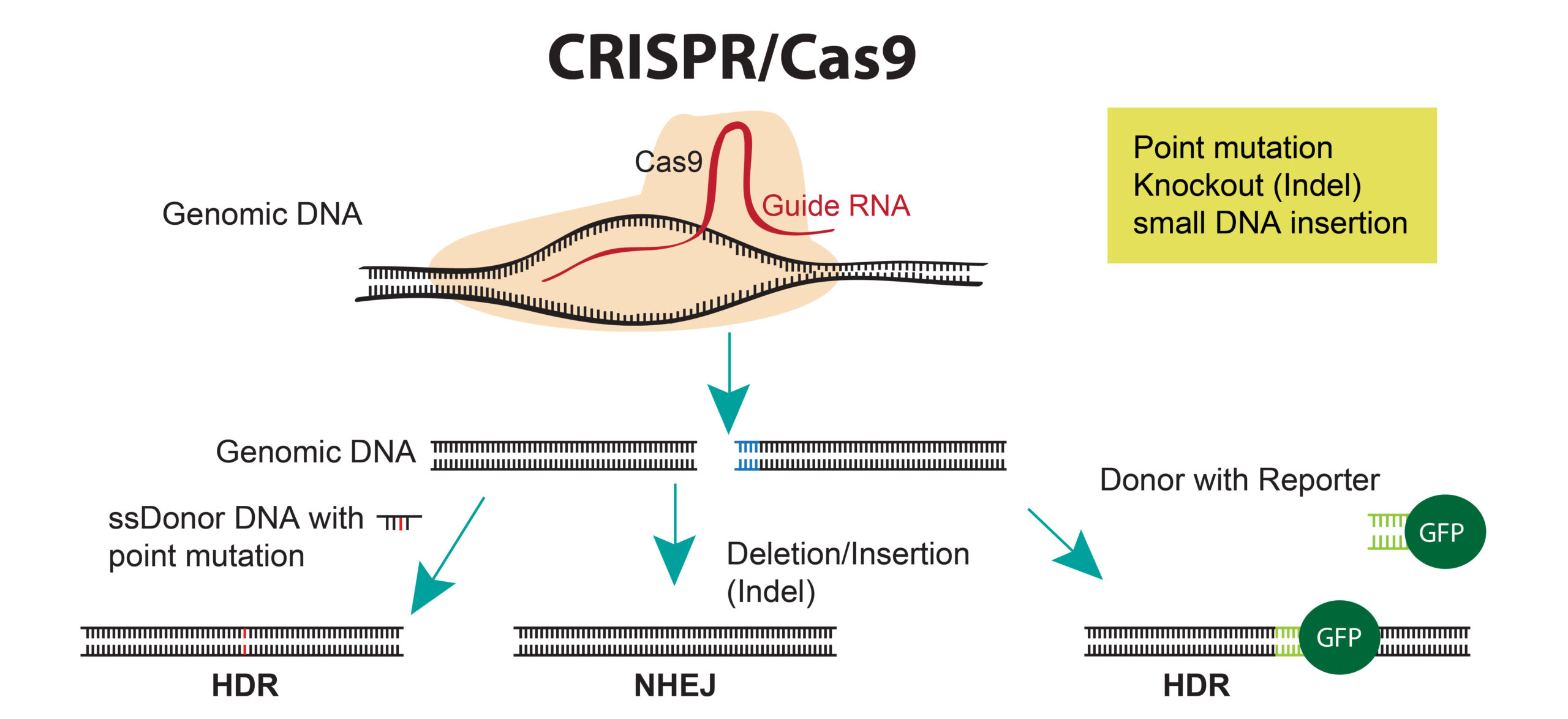
CRISPR/Cas9 is a site-specific nuclease that uses a small guide RNA to target the Cas9 nuclease to a specific sequence in the genome. Wild-type Cas9 creates double stranded breaks in the genome that can be repaired using non-homologous end joining (NHEJ)—creating an indel (insertion/deletion)—or via homologous recombination with a donor plasmid to create knock-ins or point mutations.
Challenges with CRISPR/Cas9 limit its use in therapeutic applications
How MAD7™ works
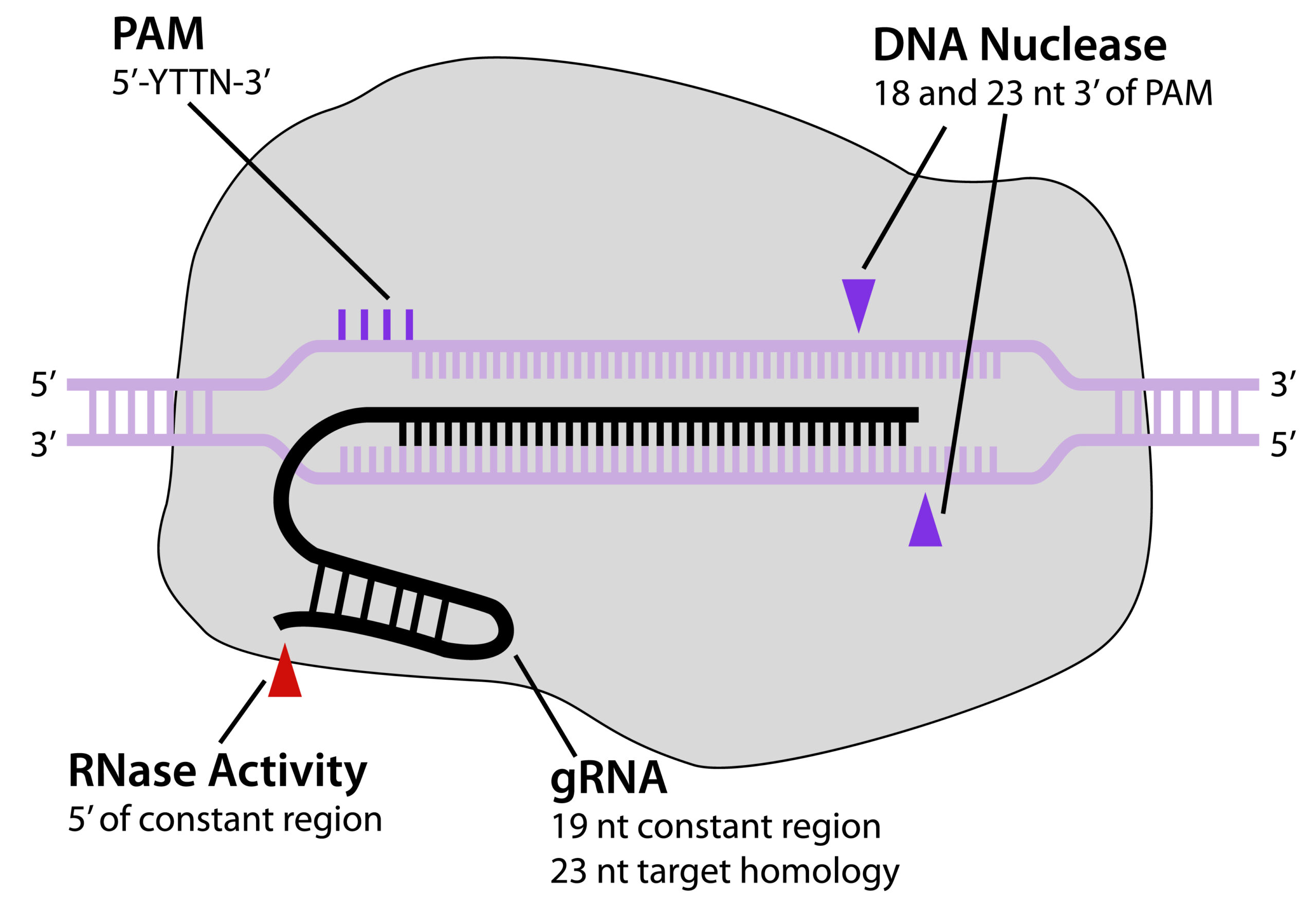
MAD7™, or ErCas12a, is a Class 2 Type V Cas nuclease that can be used for gene editing. It was developed by Inscripta and is a royalty-free alternative to CRISPR/Cas9 for academic and industrial research.
MAD7™ is based on a codon-optimized gene from Eubacterium rectale and is similar to Cas9 in that it is an RNA-guided nuclease. However, unlike Cas9, MAD7™ uses a single RNA species to guide it to the target DNA, where it creates double stranded breaks with sticky ends rather than blunt ends.
MAD7™ displays a preference for a 5′-TTTN-3′ or 5’ –CTTN-3’ PAM site rather than 5′-NGG-3′, which is preferred by Cas9.
We have successfully used MAD7™ for gene editing in iPSC lines and HEK293 cells.