From precise edits to robust expression, get iPSC genome editing done right—from the experienced innovators in genome engineering and iPSC technology
At Applied StemCell, we don’t just offer genome editing services, we specialize in editing iPSCs with the precision, consistency, and technical depth your project demands.
Our team brings years of experience in iPSC handling and genome editing, ensuring high viability, low off-target risk, and reproducible outcomes.
Whether you’re engineering a screening line, creating a disease model, or developing a therapeutic candidate, we match the iPSC genome editing technology to your project goals:
With Applied StemCell, you have a partner who understands the complexity of iPSC gene editing and delivers results you can build on.
500+
Unique iPSC lines successfully developed
>98%
Projects completed to customer specifications
1,000+
Gene editing projects completed
We support your project from start to finish, whether you only need iPSC genome editing or would like a complete iPSC workflow from reprogramming to differentiation.
As with all of our iPSC services, we can perform your iPSC gene editing service under cGMP-compliant conditions—learn more about our GMP services.
“Genome editing of iPSC cell lines was contracted through Applied Stem Cell [Milpitas, CA]. Isogenic “variant corrected” control iPSC cell lines were created for the two patient-specific LQT1 cells lines harboring KCNQ1-V254M (c.760G>A) and KCNQ1-A344A/spl (c.1032G>A).”
Single Construct Suppression and Replacement Gene Therapy for the Treatment of All CALM1-, CALM2-, and CALM3-Mediated Arrhythmia Disorders.
Hamrick SK, et al. Circ Arrhythm Electrophysiol. 2024;17(8):e012036. doi:10.1161/CIRCEP.123.012036
We match the iPSC genome editing technology to your project goals.
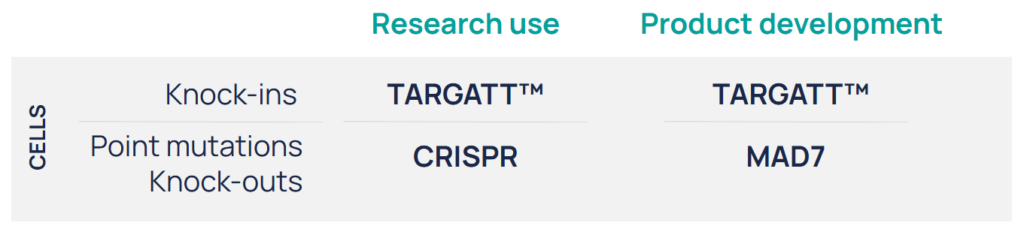
With a unique blend of iPSC expertise, genome editing mastery, and exclusive TARGATT large knock-in technology, Applied StemCell delivers unparalleled iPSC genome editing services.
We can generate the full range of genome edits, including:
“Genome correction of the SPAST mutation in iPSCs was carried out by Applied StemCell... Applied StemCell used the CRISPR/Cas9 system to correct the non-sense mutation in SPAST patients c.1684C>T/p.R562X, het. in exon 15 of the gene SPAST…Genotype of iPSCs was confirmed by the Center of Human Genetics, Regensburg University (Prof. Ute Hehr).”
Store-operated calcium entry is reduced in spastin-linked hereditary spastic paraplegia
Rizo T, et al. Brain. 2022;145(9):3131-3146. doi:10.1093/brain/awac122
Case Studies
Goal: Knock-in of 1 bp at the AAVS1 locus using the ASE-9211 Master iSPC Line by CRISPR/Cas9 technology
Knock-In Strategy for AAVS1 (1bp insertion)
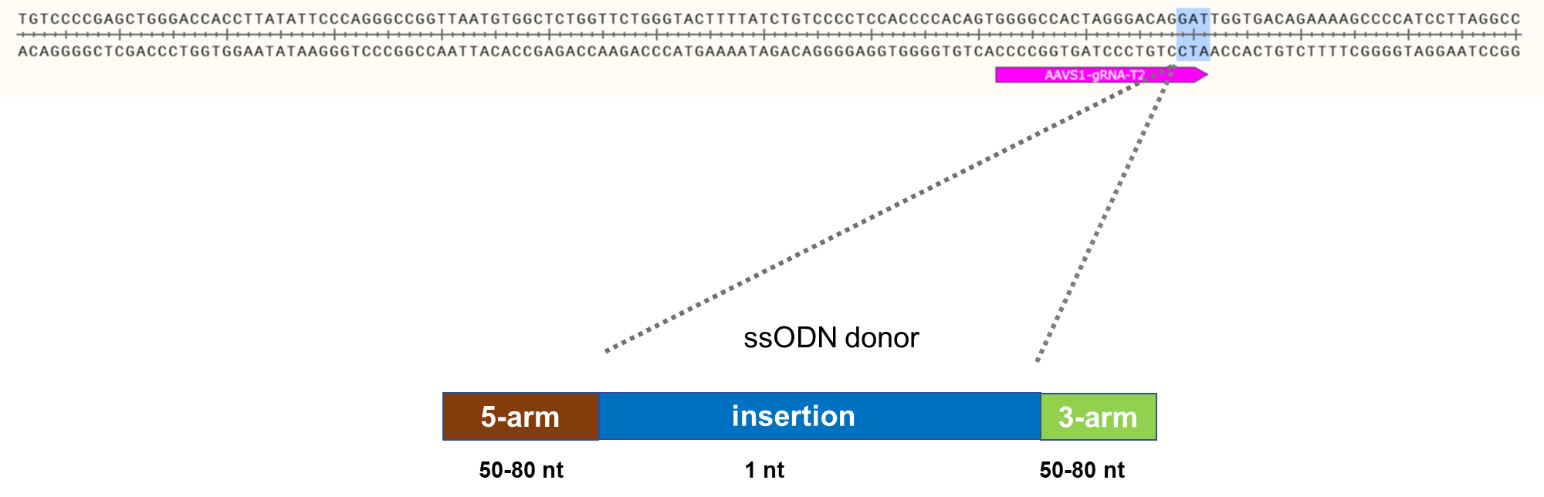
Figure 1: Knock-in strategy for 1bp insertion in the AAVS1 locus of the ASE-9211 Master Cell Line.
Genotyping Clone #6
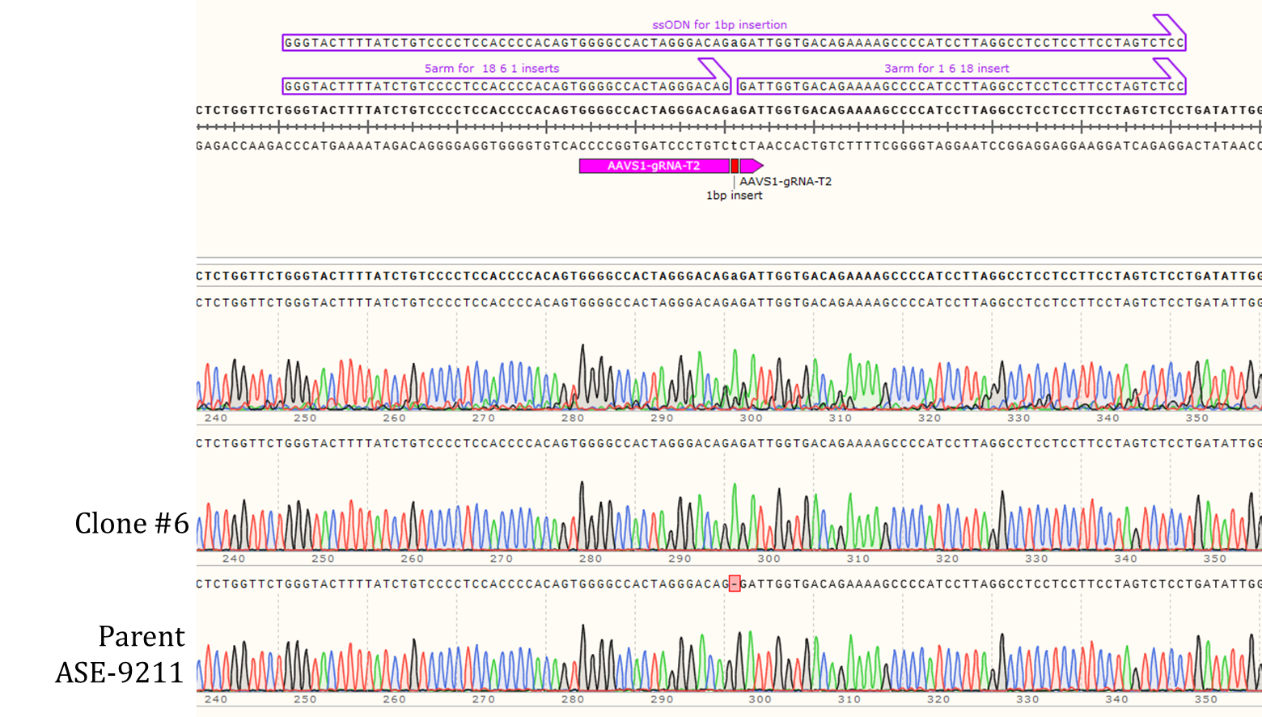
Figure 2: Sequencing chromatogram of iPSC line with 1bp insertion in the AAVS1 locus (top: Clone #6) compared to the Parent line, ASE-9211 (bottom).
Knock-In Strategy for AAVS1 (150bp insertion)
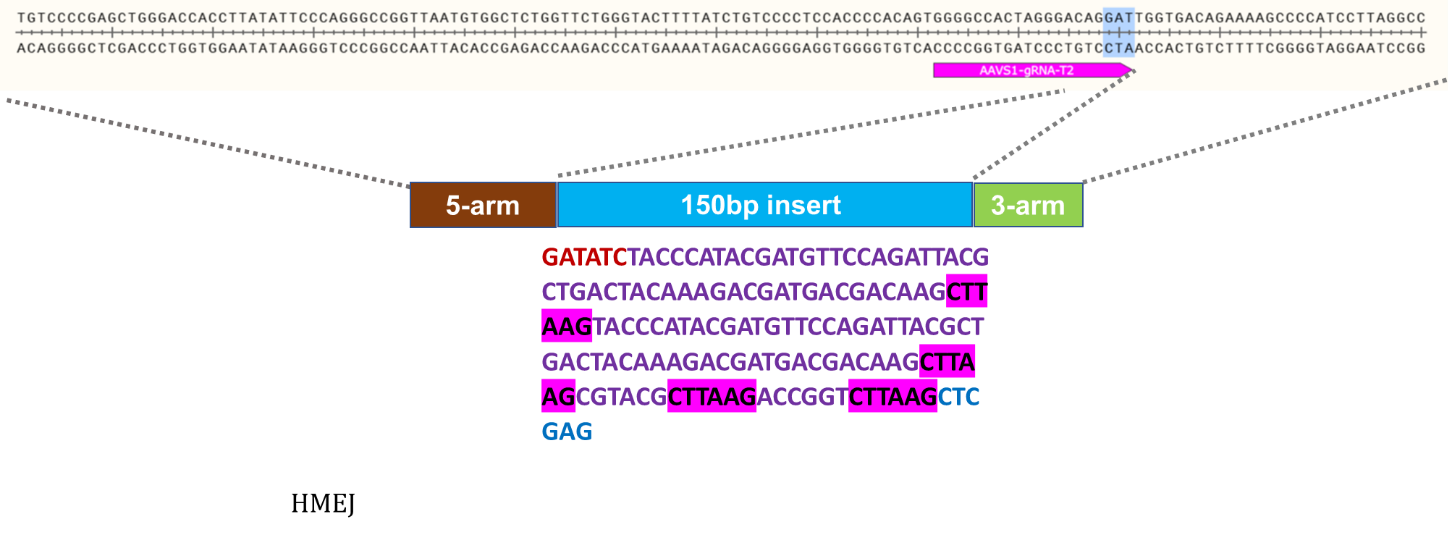
Figure 3: Knock-in strategy for 150bp insertion at the AAVS1 locus of the Master iPSC Line.
Genotyping Positive Clone #21
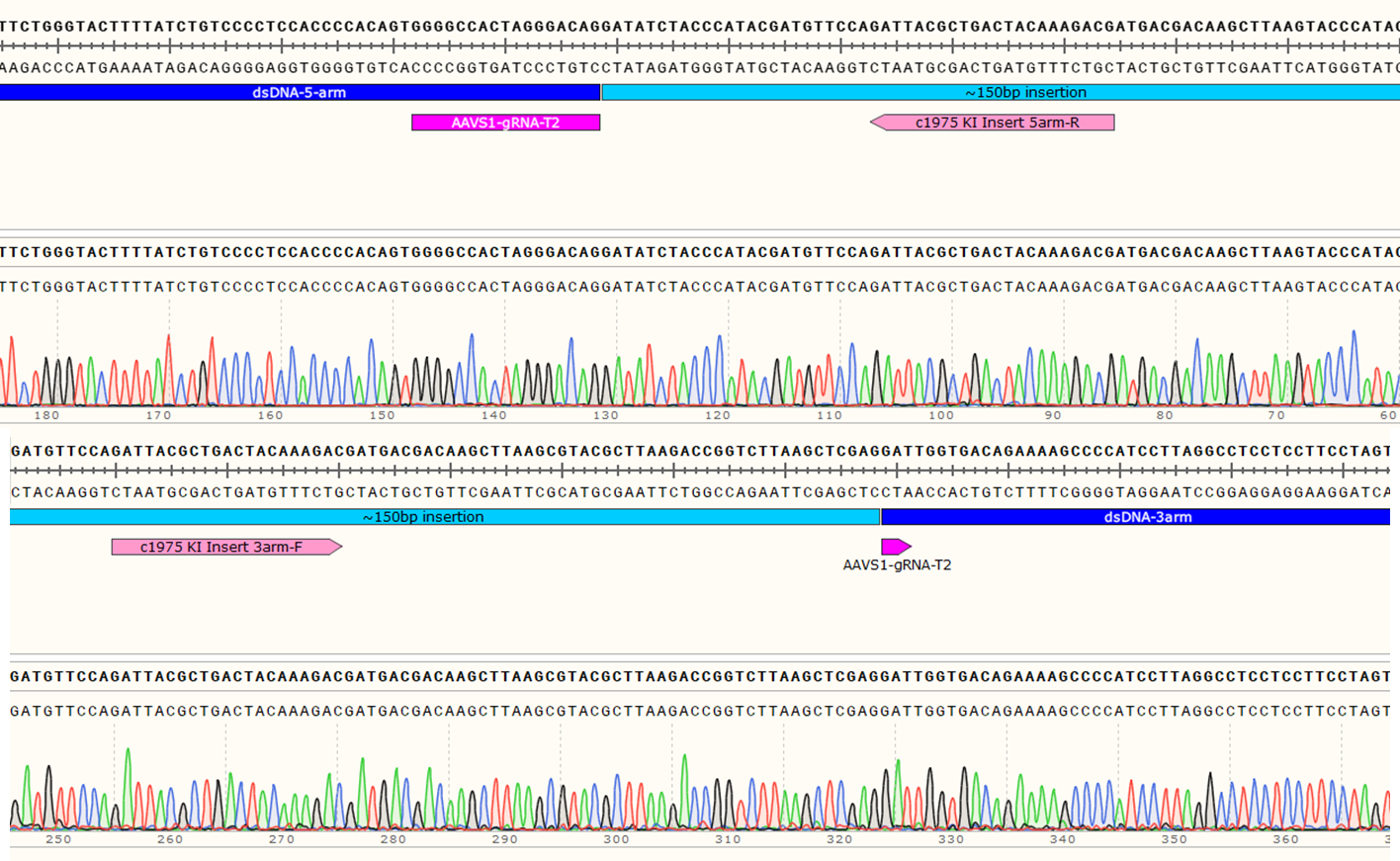
Figure 4: Sequencing chromatogram showing the ~150bp insertion at AAVS1 locus.
Goal: 1bp deletion in the AAVS1 locus using the ASE-9211 Master Cell Line by CRISPR/Cas9 technology
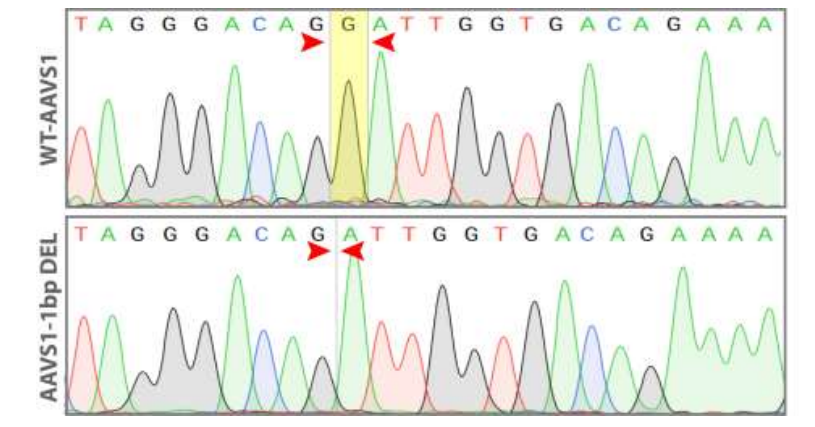
Figure 5. Sequence chromatogram of iPSC line with 1 bp deletion (AAVS1-1bp DEL; bottom) compared to wild type (WT; top).

Figure 6. Sequence alignment between the 1 bp deletion iPSC line (AAVS1-1bp DEL; bottom) and wild type (WT; top).
Goal: 1000bp Deletion in the AAVS1 locus using the ASE-9211 Master Cell Line by CRISPR/Cas9 technology
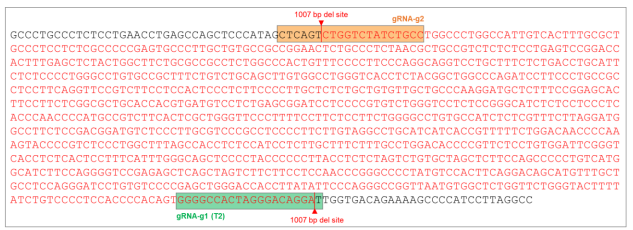
Figure 7. AAVS1 wild type (WT) sequence showing gRNA cut sites and position of 1007 bp (~1000 bp) deletion (sequence in red).
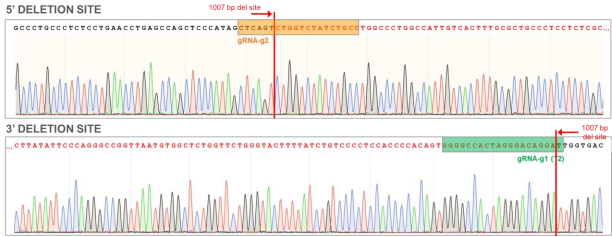
Figure 8. AAVS1 WT chromatogram showing sites of ~1000 bp deletion (sequence in red). Top: Sequence for 5’ deletion site; Bottom: Sequence for 3’ deletion site.
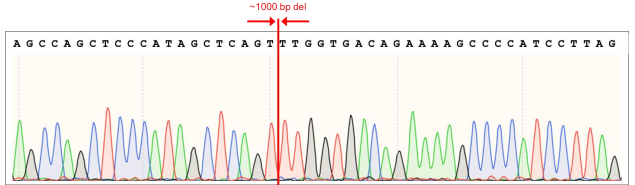
Figure 9. Sequence chromatogram of iPSC line with ~1000 bp deletion in the AAVS1 locus.
Only a few NIST projects are listed, if you would like to learn more, contact us today.
See our work in action
Single Construct Suppression and Replacement Gene Therapy for the Treatment of All CALM1-, CALM2-, and CALM3-Mediated Arrhythmia Disorders.
Hamrick SK, Kim CSJ, Tester DJ, et al. Circ Arrhythm Electrophysiol. 2024;17(8):e012036.
doi:10.1161/CIRCEP.123.012036
Characterization and AAV-mediated CRB gene augmentation in human-derived CRB1KO and CRB1KOCRB2+/− retinal organoids.
Boon N, Lu X, Andriessen CA, et al. Mol Ther Methods Clin Dev. 2023;31.
doi:10.1016/j.omtm.2023.101128
Senataxin helicase, the causal gene defect in ALS4, is a significant modifier of C9orf72 ALS G4C2 and arginine-containing dipeptide repeat toxicity.
Bennett CL, Dastidar S, Arnold FJ, et al. Acta Neuropathol Commun. 2023;11(1):164.
doi:10.1186/s40478-023-01665-z
SGK1 inhibition attenuated the action potential duration in patient- and genotype-specific re-engineered heart cells with congenital long QT syndrome.
Kim M, Das S, Tester DJ, et al. Heart Rhythm O2. 2023;4(4):268-274.
doi:10.1016/j.hroo.2023.02.003
Hereditary E200K mutation within the prion protein gene alters human iPSC derived cardiomyocyte function.
Wood AR, Foliaki ST, Groveman BR, et al. Sci Rep. 2022;12(1):15788.
doi:10.1038/s41598-022-19631-5
Store-operated calcium entry is reduced in spastin-linked hereditary spastic paraplegia.
Rizo T, Gebhardt L, Riedlberger J, et al. Brain. 2022;145(9):3131-3146.
doi:10.1093/brain/awac122
Patient-specific, re-engineered cardiomyocyte model confirms the circumstance-dependent arrhythmia risk associated with the African-specific common SCN5A polymorphism p.S1103Y: Implications for the increased sudden deaths observed in black individuals during the COVID-19 pandemic.
Hamrick SK, John Kim CS, Tester DJ, Giudicessi JR, Ackerman MJ. Heart Rhythm. 2022;19(5):822-827.
doi:10.1016/j.hrthm.2021.12.029
Coactivation of GSK3β and IGF-1 Attenuates Amyotrophic Lateral Sclerosis Nerve Fiber Cytopathies in SOD1 Mutant Patient-Derived Motor Neurons.
Ting HC, Yang HI, Harn HJ, et al. Cells. 2021;10(10):2773.
doi:10.3390/cells10102773
Dyshomeostatic modulation of Ca2+-activated K+ channels in a human neuronal model of KCNQ2 encephalopathy.
Simkin D, Marshall KA, Vanoye CG, et al. eLife. 2021;10:e64434.
doi:10.7554/eLife.64434
Suppression-Replacement KCNQ1 Gene Therapy for Type 1 Long QT Syndrome.
Dotzler SM, Kim CSJ, Gendron WAC, et al. Circulation. 2021;143(14):1411-1425. doi:10.1161/CIRCULATIONAHA.120.051836
Find out how our iPSC genome editing services can enable your projects by submitting the form below. We will get back to you within one business day.