Accelerate cell line development for library creation and protein expression with efficient TARGATT™ large knock-in technology
From promoter screening libraries to protein expression and stable viral vector production, TARGATT™ HEK293 Kits put efficient and precise cell line development into your hands.
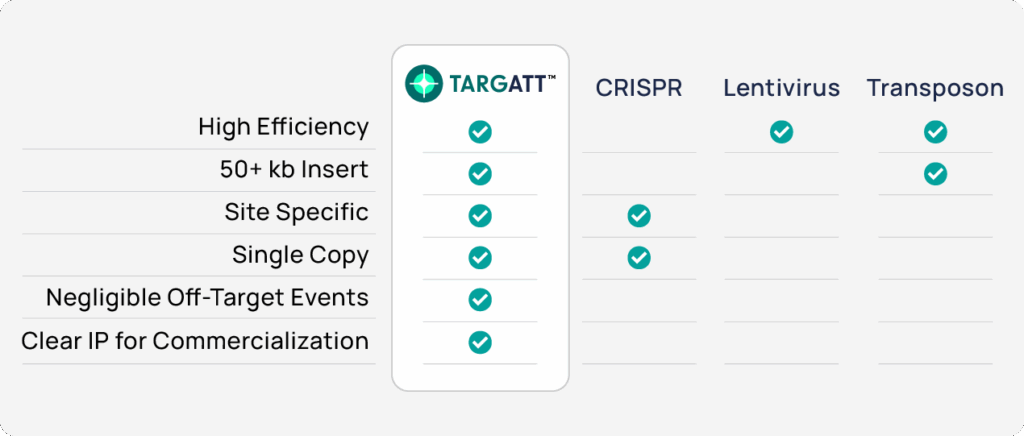
Leveraging the power of site-specific integration and the consistently-expressing H11 safe harbor site, our TARGATT™ large knock-in technology enables genome engineering that CRISPR, transposons, and lentivirus can’t handle.
TARGATT™ Kits are designed to enable easy evaluation of the technology before purchasing a license*. Each TARGATT™ HEK293 Kit includes a HEK293 cell line with the TARGATT™ landing pad already inserted into the H11 safe harbor site, a donor plasmid for the DNA you are knocking in, a positive control plasmid, and a TARGATT™ Integrase expression plasmid.
*Academic researchers may purchase TARGATT™ technology without licensing. Request more information.
If you are interested in outsourcing genome engineering, contact us for a custom service project quote.
Learn how TARGATT™ technology works and see the data—visit the technology page.
Because integration using TARGATT™ technology is so efficient, you can create highly diverse libraries and even perform mammalian display for screening in a biologically-relevant cell type.
A high throughput bispecific antibody discovery pipeline
Segaliny AI, Jayaraman J, Chen X, et al.
Communications Biology. 2023;6(1). doi:10.1038/s42003-023-04746-w
Fill out the contact form below and a team member will be in touch within one business day.
Publications about TARGATT™ technology