Build cell-based models of disease and accelerate therapeutic development with TARGATT™ large knock-in technology in iPSCs
Whether you are creating tomorrow’s iPSC-based treatments or building a cellular model of disease, TARGATT™ iPSC kits will simplify and accelerate cell line development when you need to knock-in and express one or more genes.
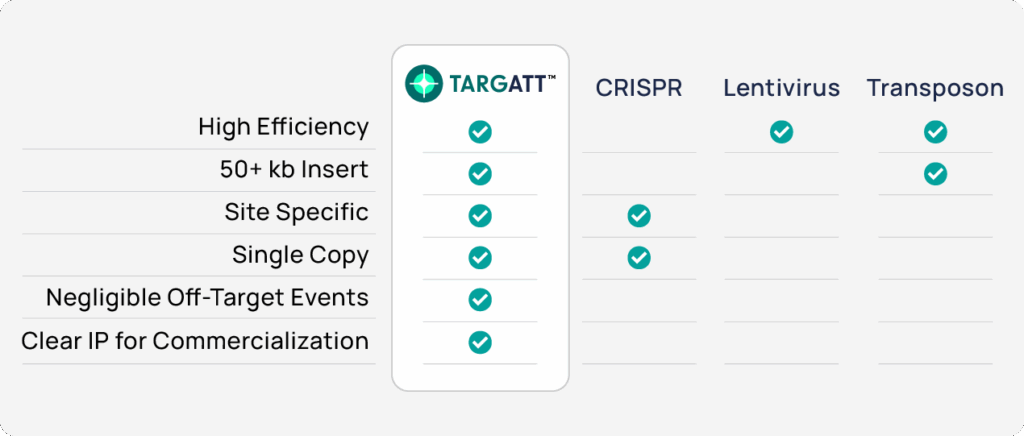
Combining the power and safety of our TARGATT™ large knock-in technology with the scalability and isogenic consistency of iPSCs, TARGATT™ iPSC Kits enable genome engineering that CRISPR, transposons, and lentivirus can’t handle.
TARGATT™ Kits are designed to enable easy evaluation of the technology before purchasing a license.* Each TARGATT™ iPSC kit includes a human iPS cell line with the TARGATT™ landing pad already inserted into the H11 safe harbor site, a donor plasmid for the DNA you are knocking in, and a TARGATT™ Integrase expression plasmid.
TARGATT™ kits are available with iPSCs derived from male CD34+ umbilical cord blood cells (AST-9450, AST-9480, AST-9650) as well as iPSCs derived from fibroblasts from an adult female donor (AST-1602).
Contact us if you wish to purchase additional kit components.
*Academic researchers may purchase TARGATT™ technology without licensing. Request more information.
If you are interested in outsourcing genome engineering and/or differentiation, contact us for a custom service project quote.
Learn how TARGATT™ technology works and see the data—visit the technology page.
Fill out the contact form below and a team member will be in touch within one business day.
Publications about TARGATT™ technology